Intramolecular dialogue within the human proteasome
The proteasome is a proteolytic machinery allowing the recycling of intracellular proteins. Its deregulation has been associated with various neurodegenerative diseases and certain cancers. This work, published in the journal Nature Communications, reveals a remote enzymatic regulation mechanism between the catalytic sites buried in the proteasome and the surface accessible to its activators. The method used opens up prospects for the study of many other modulators of its activity and other multi-protein complexes.
The proteasome is a multi-protein complex responsible for degrading certain proteins and thus maintaining protein homeostasis. Its activity is also responsible for the intracellular concentration of numerous cytokines and other « hub » proteins involved in cellular processes such as cell division, inflammation or the generation of antigenic peptides. It is therefore subject to very fine regulation depending on cellular needs and physiological or external disturbances.
This proteolytic activity can be modulated by interacting with different regulators, by replacing catalytic subunits or by post-translational modification. The literature lists more than 300 proteins interacting with the catalytic core of the proteasome. However, the catalytic sites of the proteasome are buried at the heart of this vast multi-protein assembly (28 subunits assembled in the form of 4 rings made up of 7 different subunits), far from the surface accessible to possible regulatory molecules or proteins. The molecular mechanisms explaining this remote regulation are still poorly understood.
This study has made it possible to better describe these intramolecular dialogues regulating proteasome activity, by adapting an innovative methodology to this gigantic multi-protein complex. This technique, based on hydrogen-deuterium exchange, makes it possible to estimate the accessibility to the solvent of the different regions of a protein.
Therefore, by comparing the data obtained on proteasomes containing different catalytic subunits (standard proteasomes and immunoproteasomes), the researchers have highlighted fine intramolecular perturbations starting from these subunits and gradually reaching the surface of the complex. These results explain how the introduction of this or that isoform into the complex can promote interaction with this or that specific regulator. Moreover, the comparison of these proteasomes with or without regulators has made it possible to highlight a reverse regulation mechanism, in which the contact at the proteasome surface is propagated to the catalytic sites.
The successful use of this methodology based on hydrogen deuterium-exchange on a complex the size of the proteasome shows that it can undoubtedly be applied to other supramolecular systems (ribosomes, inflammasomes…).

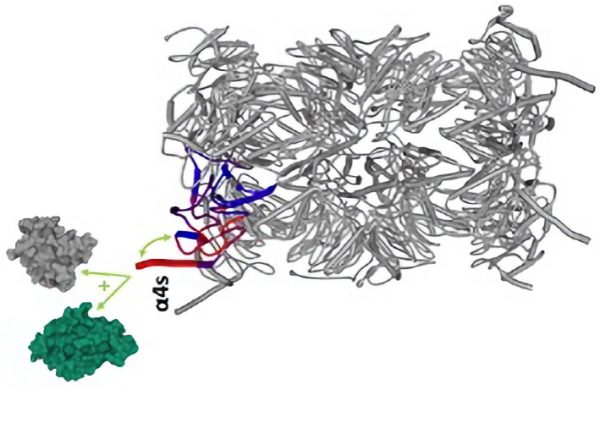
Figure: Representation of the 3D structure of the standard human proteasome (PDB: 5LE5) highlighting regions that are more (red) or less (blue) flexible/accessible after interaction with the PA28αβ activator. These results of hydrogen-deuterium exchange coupled with mass spectrometry make it possible to visualise the areas affected during the formation of the complex.