Large–scale proteomic profiling of regulatory T cells
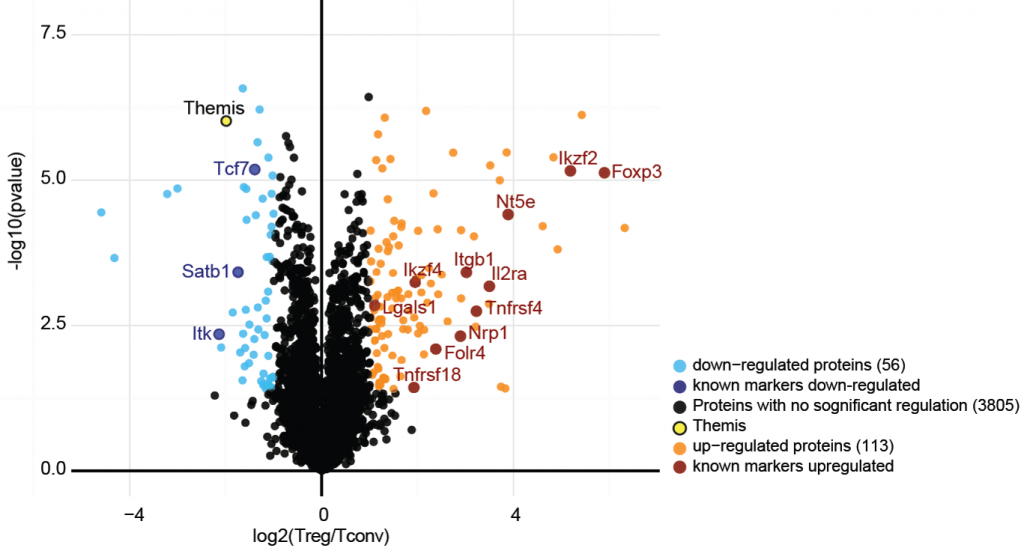
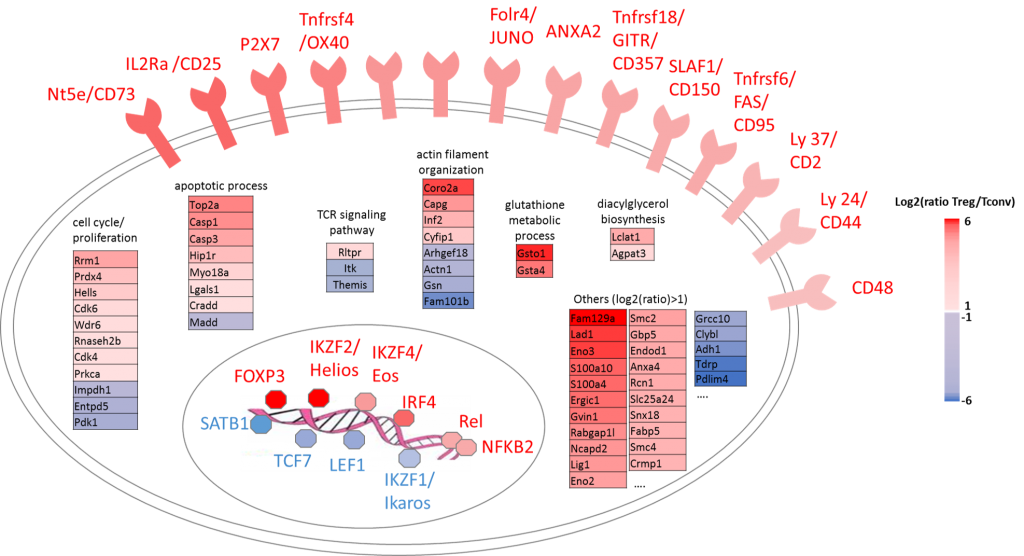
Regulatory T cells (Treg) represent a minor sub-population of T lymphocytes which is crucial for the maintenance of immune homeostasis. Using large-scale quantitative mass spectrometry, we performed an in-depth comparison of the proteomic profile of Treg versus conventional T cells, and defined a specific proteomic “signature” of Treg (1).
We focused on Themis1, a protein particularly under-represented in Treg, and described as being involved in the pathogenesis of immune diseases. Using a transgenic mouse model over-expressing Themis1, we provided in vivo and in vitro evidence of its importance for Treg suppressive functions. We also characterized the interactome of Themis1 in primary mouse thymocytes, through immunopurification of endogenous Themis1 (2).
1- Duguet F, Locard-Paulet M et al. Proteomic analysis of regulatory T cells reveals the importance of Themis1 in the control of their suppressive function. Mol Cell Proteomics. 2017.
https://doi.org/10.1074/mcp.m116.062745
2- Zvezdova et al (2016) Themis1 enhances T cell receptor signaling during thymocyte development by promoting Vav1 activity and Grb2 stability. Sci Signal, 2016.
Proteomic signature of regulatory T cells. Large-scale proteomics allows measuring cellular protein abundaces at high-throughput. By comparing Treg with conventional T cells, a panel of marker proteins specifically up-or down-regulated in this population were identified.
