Functional mechanisms of interleukin-33 in allergic inflammation
In collaboration with the team of J-P Girard at IPBS, we use proteomic analysis associated with label-free quantification to study inflammatory mechanisms, particularly the mode of action of Interleukin-33 (IL-33), a nuclear cytokine involved in type 2 immunity, inflammation and allergy.
More than 10 years ago, we developed original bioinformatics tools to extract peptide signals from raw MS files, measure their intensity, and perform quantitative analysis of protein expression in large-scale studies. Using such methods, we could extensively map at that time the response of human endothelial cells to several inflammatory cytokines, such as TNFα, IFNg and IL1β (1,2).
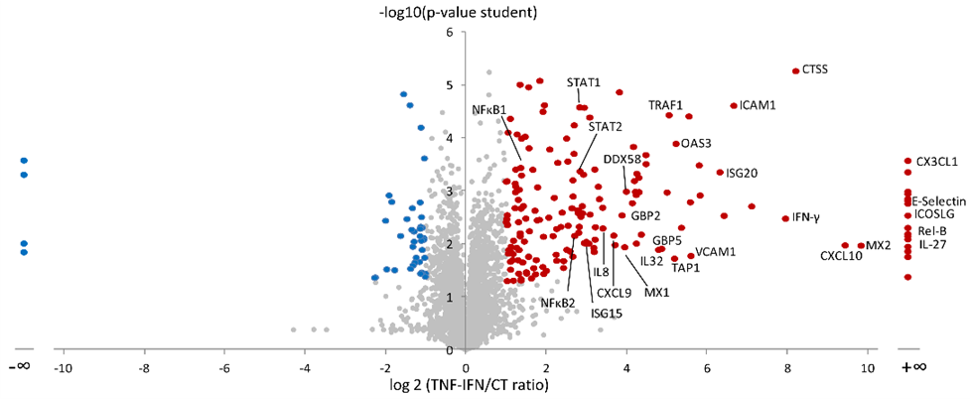
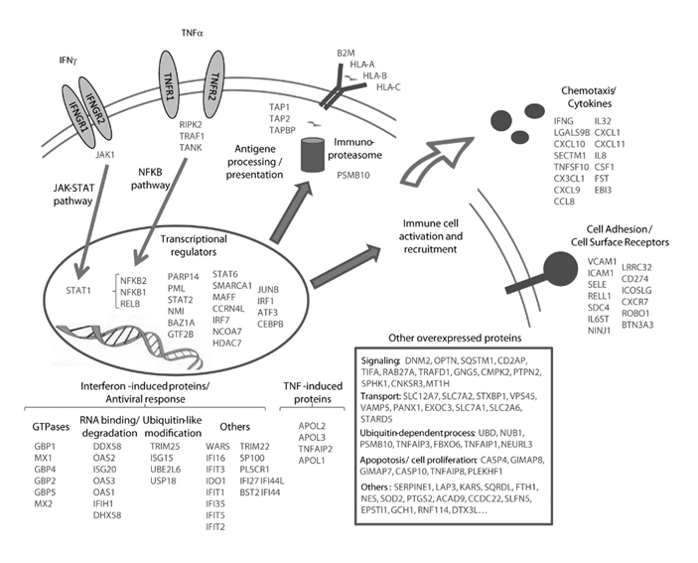
We keep on applying such a global proteomic approaches to analyze IL-33 function in endothelial cells and other target immune cells such as group 2 innate lymphoid cells (ILC2s), which play an important role in asthma.
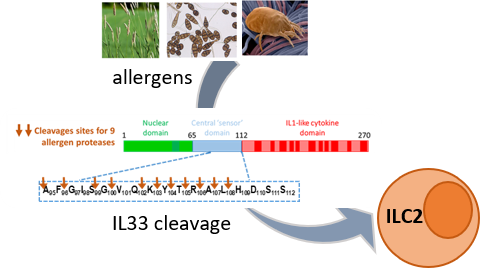
We use large-scale protein profiling to compare the extracellular and intracellular activity of IL-33 in primary human endothelial cells (3). Using highly sensitive mass spectrometry, we could also identify the cleavage sites generated on IL-33 by a whole panel of allergens. These cleavage events induce activation of the cytokine, finally triggering asthma (4,5).
- Bouyssie, Gonzalez de Peredo et al. Mascot file parsing and quantification (MFPaQ), a new software to parse, validate, and quantify proteomics data generated by ICAT and SILAC mass spectrometric analyses: application to the proteomics study of membrane proteins from primary human endothelial cells. Molecular & cellular proteomics , 2007 https://doi.org/10.1074/mcp.t600069-mcp200
- Gautier et al. Label-free quantification and shotgun analysis of complex proteomes by one-dimensional SDS-PAGE/NanoLC-MS: evaluation for the large scale analysis of inflammatory human endothelial cells. Molecular & cellular proteomics, 2012 https://doi.org/10.1074/mcp.m111.015230
- Gautier et al. Extracellular IL-33 cytokine, but not endogenous nuclear IL-33, regulates protein expression in endothelial cells. Sci Rep, 2016 https://doi.org/10.1038/srep34255
- Lefrancais et al. IL-33 is processed into mature bioactive forms by neutrophil elastase and cathepsin G. Proc Natl Acad Sci U S A 2012. https://doi.org/10.1073/pnas.1115884109
- Cayrol et al. Environmental allergens induce allergic inflammation through proteolytic maturation of IL-33. Nat. Immunol. 2018. https://doi.org/10.1038/s41590-018-0067-5
