
Hydrogen-deuterium exchange enables to detect conformational differences between two protein samples.
The proteins or protein complexes are diluted in a deuterated buffer and hydrogen atoms from the peptide bonds are exchanged with the deuterium atoms of the solution. The rate of exchange of each amide hydrogen depends on its solvent accessibility and involvement in the stabilization of secondary structures. By determining the rate of deuterium exchange, we can identify which regions are solvent-accessible and which ones are hidden in the core of the protein.
This expanding method can thus be used to study protein dynamics, compare protein conformations (in different buffer conditions, after mutation), or to identify protein-protein or protein-ligand interfaces.
 Figure 1. Automated HDX-MS solution including the sample preparation robot (LeapTec), the M-Class Acquity UPLC, the HDX-module for protein digestion and peptide separation at 4°C and the SynaptG2Si (Waters).
Figure 1. Automated HDX-MS solution including the sample preparation robot (LeapTec), the M-Class Acquity UPLC, the HDX-module for protein digestion and peptide separation at 4°C and the SynaptG2Si (Waters).
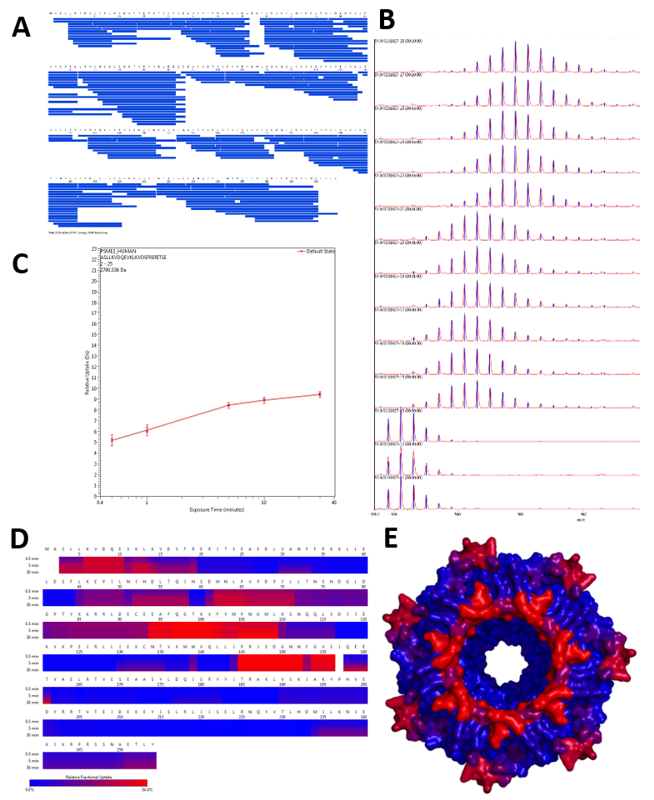 Figure 2. Dynamix (Waters) software automatically generates sequence coverage maps (A), extracts the mass increment of each peptide at each timepoint (B) to draw deuteration curve (C). Heatmaps are then generated (D) and can be imported to Pymol, if a structure of the protein of interest is available (E).
Figure 2. Dynamix (Waters) software automatically generates sequence coverage maps (A), extracts the mass increment of each peptide at each timepoint (B) to draw deuteration curve (C). Heatmaps are then generated (D) and can be imported to Pymol, if a structure of the protein of interest is available (E).2023
Yang Y, Chen H#, Corey R#, Morales V#, Quentin Y, Froment C, Caumont-Sarcos A, Albenne C, Burlet-Schiltz O, Ranava D, Stansfeld P, Marcoux J, Ieva R. LptM promotes oxidative maturation of the lipopolysaccharide translocon by substrate binding mimicry. Nature Communications. 14(1):6368.
2022
Živković D, Sanchez Dafun A, Menneteau T, Schahl A, Lise S, Kervarrec C, Toste Rêgo A, da Fonseca PCA, Chavent M, Pineau C, Burlet-Schiltz O, Marcoux J, Bousquet MP. Proteasome complexes experience profound structural and functional rearrangements throughout mammalian spermatogenesis. Proceedings of the National Academy of Sciences. 119(15):e2116826119.
Chaptal V, Zampieri V, Wiseman B, Orelle C, Martin J, Nguyen KA, Gobet A, Di Cesare M, Magnard S, Javed W, Eid J, Kilburg A, Peuchmaur M, Marcoux J, Monticelli L, Högbom M, Schoehn G, Jault JM, Boumendjel A, Falson P. Substrate-bound and substrate-free outward-facing structures of a multidrug ABC exporter. Science Advances. 8(4):eabg9215.
Javed W, Vallet S, Clement MP, Le Roy A, Moulin M, Härtlein M, Breyton C, Burlet-Schiltz O, Marcoux J, Orelle C, Ebel C, Martel A, Jault JM. Structural insights into the catalytic cycle of a bacterial multidrug ABC efflux pump. J Mol Biol. 167541.
2020
Lesne J, Locard-Paulet M, Parra J, Zivković D, Menneteau T, Bousquet MP, Burlet-Schiltz O, Marcoux.Conformational maps of human 20S proteasomes reveal PA28- and immuno-dependant inter-ring crosstalks. Nature Communications 11(1):6140.
2019
Guillet V, Bordes P, Bon C, Marcoux J, Gervais V, Sala AJ, Dos Reis S, Slama N, Mares-Mejía I, Cirinesi AM, Maveyraud L, Genevaux P, Mourey L. Structural insights into chaperone addiction of toxin-antitoxin systems. Nature Communications 10(1):782.
Bouyssié D, Lesne J, Locard-Paulet M, Albigot R, Schiltz O, Marcoux J. HDX-Viewer: interactive 3D visualization of Hydrogen-Deuterium eXchange data. BioInformatics 35(24):5331-5333.
